2. 华东师范大学 地理信息科学教育部重点实验室, 上海 200241;
3. 华东师范大学 地理科学学院, 上海 200241
2. Key Laboratory of Geographic Information Science(Ministry of Education), East China Normal University, Shanghai 200241, China;
3. School of Geographic Sciences, East China Normal University, Shanghai 200241, China
近年来, 人为来源活性氮的过量排放严重干扰了河口生态系统的氮循环过程, 由此引发了水体富营养化等系列环境问题.因此, 河口近岸生态系统氮循环过程及其功能微生物类群成为广泛关注的研究热点.硝化过程(将氨氮(NH
目前, 有关自然环境中AOB与AOA的菌群多样性、空间分布差异等方面的研究较多[5-7], 而NOB的研究相对滞后.一方面, 因为亚硝酸盐氧化不是硝化过程的限速步骤, 所以亚硝酸盐氧化过程的关注度较低; 另一方面, 因为NOB Nitrospira属难培养, 发现新种系的可能较小, 所以NOB的相关研究也较少[8].而comammox的发现更晚于AOB、AOA和NOB类群, 并迅速成为氮循环过程的研究热点.首次报道的三株comammox(Candidatus Nitrospira inopinata, Candidatus Nitrospira nitrosa和Candidatus Nitrospira nitrificans)均发现于工业系统[3-4], 之后在饮用水系统、活性污泥、森林土壤和湖泊沉积物等生态系统都检测到comammox的存在[3-4, 9-16].
河口近岸区域是地球上最多产的生态系统之一, 是生物地球化学循环的重要组成部分, 是氮循环研究的热点区域[17-18].目前, 在河口潮滩生态系统中对硝化功能微生物还缺乏深入认识, 尤其是新发现的comammox类群, 其生态位及与其他硝化功能菌的相互作用还不甚清楚.受潮汐变化、盐水入侵以及脉冲式营养盐输入等海陆交互作用的影响, 河口沉积环境微生物群落在系统分类和功能代谢上较为复杂[19].富集培养是微生物探究的重要方法, 可以放大研究目标, 简化研究体系[20].为获得河口潮滩生态系统具有较高硝化能力的微生物群体, 提供后续硝化微生物代谢和影响机制探究的实验基础, 简化河口潮滩复杂环境因子的影响, 本研究进行了硝化微生物的富集培养.在此基础上, 结合宏基因组和宏转录组技术, 从DNA和mRNA两个层面对硝化富集培养物进行了种群结构和基因表达活性的分析.研究工作深化了对河口潮滩氮素生物地球化学循环的理论认识, 对于深入开展河口环境硝化过程的分子生态学研究有重要借鉴.
1 材料与方法 1.1 沉积物的富集培养为了避免复杂环境因子的干扰, 更具体地分析河口潮滩地区的硝化过程, 选择氨氧化(硝化过程的限速步骤)细菌的最适生长条件, 进行了硝化微生物的富集培养.将4.0 L工作容积的生物反应器湿热灭菌后, 接种50 g新鲜的长江口潮滩沉积物, 并配备pH、温度、溶解氧和液面高度监测的探针.设置培养基的加入流速1.4 mL/min, 搅拌速度为每分钟250转.通过加入100 g/L的KHCO
为分析硝化菌群
经一个月的富集培养后, 使用PowerSoil®DNA Isolation Kit (MO BIO, USA)提取3份反应器富集物的总DNA.将3份DNA样品混匀, 使用Covaris S220 Focused Ultrasonicator将总DNA随机打断成350 bp的片段.通过PCR反应对这些片段进行末端修复, 连接poly-A尾并添加接头构建测序文库, 基于Illumina Hiseq平台, 进行150-bp双末端测序.得到的原始数据进入信息分析流程.首先去除原始测序reads的接头, 去除threshold quality
| $ \begin{align*} G_{k }=\frac{r_k}{L_k}\cdot\frac{1}{\sum^n_{i=1}\frac{r_i}{L_i}}. \end{align*} $ |
上式中,
经一个月的富集培养后, 使用EZNA®Soil RNA kit (Omega Bio-tek, Norcross, GA, USA)试剂提取三份反应器富集物的RNA.使用Ribo-Zero Magnetic Kit (Epicenter, Charlotte, NC, USA)去除rRNA, 混合三份样品, 使用TruSeq
一个月的培养过程中, 反应器体系各理化因子测量值变化趋势如图 1.培养基的NH
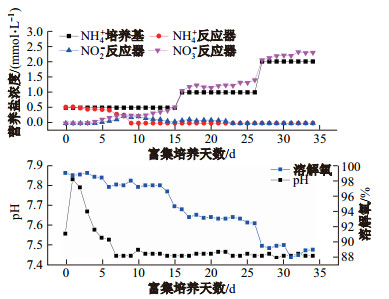
|
图 1 培养基及反应器中NH |
然而,
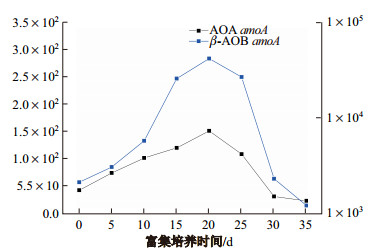
|
注:图中左侧纵坐标表示每ng DNA中AOAamoA的基因拷贝数, 右侧是每ng DNA中 |
氨氧化微生物的丰度变化与反应器中硝化能力增强的趋势并不一致, 暗示反应体系中可能存在进行氨氧化作用的非传统微生物.与此同时, 新近comammox的发现指明反应体系中可能存在该类群微生物.
2.2 宏基因组数据表征富集培养物的菌群丰度对富集物总DNA测序获得20 Gb数据, 构建的非冗余基因集包含762, 516个ORFs, 其平均长度为650.40 bp.统计获得菌群丰度结果如图 3, 图中显示了硝化类群(
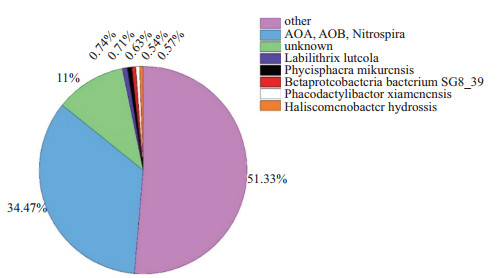
|
图 3 生物反应器中微生物类群丰度 Fig.3 The abundance of microorganisms in the bioreactor system |
图 4显示硝化微生物群体的具体组成, 图中内圈扇形表示富集物在种水平上注释到的硝化菌群的丰度, 外圈表示富集物中4类硝化菌群comammox, NOB,
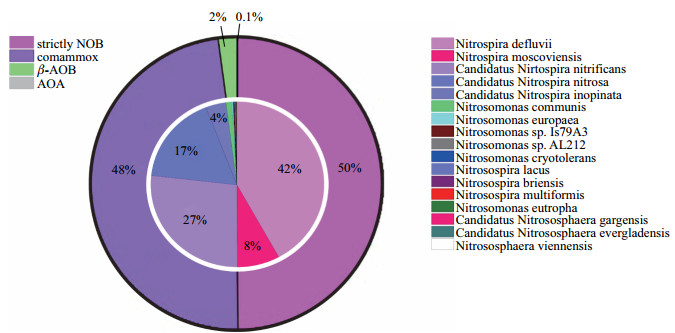
|
注:各类群丰度为该类群占硝化菌群的比例.图中内圈表示富集物在种水平上注释到的主要硝化菌群的丰度(右侧图例), 外圈表示富集物中4类硝化菌群的丰度(左侧图例) 图 4 生物反应器中硝化菌群丰度 Fig.4 The abundance of nitrifiers in the bioreactor system |
宏基因组结果从种群结构角度揭示了富集的硝化微生物中comammox和NOB的优势地位. AOB、AOA和comammox三类菌群的竞争关系源于它们都通过对底物NH
宏基因组结果呈现了富集物的种群结构, 揭示了非传统氨氧化微生物——comammox的主导地位.结果暗示在长江河口潮滩沉积物中, 存在comammox菌群.硝化微生物的注释结果中,
生物反应器富集培养过程中, 设定培养条件(pH, 溶氧, 温度)为氨氧化(硝化过程的限速步骤)细菌的最适生长条件, 使培养基中NH
对富集物总RNA反转录测序后获得14 Gb数据, 预测得到43 417个ORFs.比对eggNOG数据库将功能基因分为25个功能大类, 富集培养物4类硝化菌群的基因表达量在25个功能大类分布情况的统计结果如图 5. Comammox的代谢中, 翻译后修饰、蛋白质转换相关的基因表达量最高.能量生产及转化, 细胞壁、细胞膜合成, 信号传导以及翻译、核糖体构建、生物合成的基因表达次之, 暗示了富集物中comammox非常高的代谢活性和细菌增殖趋势. NOB基因表达量的高低分布情况与comammox相近, 但略低于comammox. AOA代谢中, 防御机制相关的基因表达量最高, 且远高于其他3个菌群, 翻译、核糖体构建、生物合成等代谢次之.防御机制相关基因表达量高的现象可能是生物反应器选择了AOB的富集培养条件(与富集培养AOA的pH、培养基成分等条件不同[40-41]), 不适合的培养环境刺激产生了防御机制.氨氧化细菌在翻译, 核糖体构建, 生物合成和无机离子传输、代谢方面基因表达量较高.氨氧化微生物类群细胞壁、细胞膜合成代谢的基因表达量非常低, 暗示富集物中氨氧化微生物较小的增殖空间.
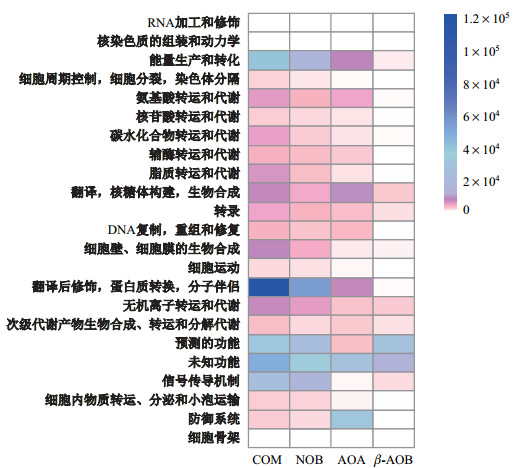
|
注:图中NOB指严格亚硝酸盐氧化的NOB, COM指全程氨氧化微生物, AOA指氨氧化古菌, |
4类菌群预测性的和未知功能的基因表达量都较高, comammox的最高, 暗示菌群代谢中较多的知识空白.总体上, 4类菌群中, comammox基因表达量略高于NOB, 二者远高于氨氧化微生物.宏基因组数据中comammox与NOB的高丰度优势与宏转录组中二者的高表达量优势一致.结果表明本研究培养条件下, comammox和NOB具有较高的代谢活性, 与comammox在极贫营养条件下更有竞争优势的特点[42-43]一致. Nitrospira在实验室难培养[8, 44]的特点暗示本研究的富集培养对于Nitropira属和comammox的研究具有重要意义.
3 结论本研究采集河口潮滩沉积物, 进行硝化微生物的富集培养.宏基因组结果揭示了硝化微生物的种群结构, 主要包括4类硝化菌群——comammox, NOB,
| [1] |
LAM P, LAVIK G, JENSEN M M, et al. Revising the nitrogen cycle in the Peruvian oxygen minimum zone[J]. Proc Natl Acad Sci USA, 2009, 12(106): 4752-4757. |
| [2] |
KRAFT B, HALINA E T, RITIN S, et al. The environmental controls that govern the end product of bacterial nitrate respiration[J]. Science, 2014, 345(6197): 676-679. DOI:10.1126/science.1254070 |
| [3] |
VAN KESSEL M A, SPETH D R, ALBERTSEN M, et al. Complete nitrification by a single microorganism[J]. Nature, 2015, 528(7583): 555-559. DOI:10.1038/nature16459 |
| [4] |
DAIMS H, LEBEDEVA E V, PJEVAC P, et al. Complete nitrification by Nitrospira bacteria[J]. Nature, 2015, 528(7583): 504-509. DOI:10.1038/nature16461 |
| [5] |
LI J, NEDWELL D B, BEDDOW J, et al. AmoA gene abundances and nitrification potential rates suggest that benthic ammonia-oxidizing bacteria and not archaea dominate N cycling in the Colne Estuary, United Kingdom[J]. Appl Environ Microbiol, 2014, 81(1): 159-165. |
| [6] |
LAGOSTINA L, GOLDHAMMER T, RØY H, et al. Ammonia-oxidizing bacteria of the Nitrosospira cluster 1 dominate over ammonia-oxidizing archaea in oligotrophic surface sediments near the South Atlantic Gyre[J]. Environ Microbiol Reports, 2015, 7(3): 404-413. DOI:10.1111/emi4.2015.7.issue-3 |
| [7] |
SHEN J, ZHANG L, DI H J, et al. A review of ammonia-oxidizing bacteria and archaea in Chinese soils[J]. Frontiers in Microbiology, 2012(3): 296. |
| [8] |
DAIMS H, LÜCKER S, WAGNER M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria[J]. Trends in Microbiology, 2016, 24(9): 699-712. DOI:10.1016/j.tim.2016.05.004 |
| [9] |
PINTO A J, MARCUS D N, IJAZ U Z, et al. Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system[J]. Msphere, 2015(1): e15-e54. |
| [10] |
CHAO Y, MAO Y, YU K, et al. Novel nitrifiers and comammox in a full-scale hybrid biofilm and activated sludge reactor revealed by metagenomic approach[J]. Appl Microbiol Biotechnol, 2016, 100(18): 8225-8237. DOI:10.1007/s00253-016-7655-9 |
| [11] |
PALOMO A, JANE FOWLER S, GÜLAY A, et al. Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp.[J]. The ISME Journal, 2016, 10(11): 2569-2581. DOI:10.1038/ismej.2016.63 |
| [12] |
WANG Y, MA L, MAO Y, et al. Comammox in drinking water systems[J]. Water Research, 2017, 116: 332-341. DOI:10.1016/j.watres.2017.03.042 |
| [13] |
BARTELME R P, MCLELLAN S L, NEWTON R J. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira[J]. Frontiers in Microbiology, 2017(8): 101. |
| [14] |
WANG J, XIA F, ZELEKE J, et al. An improved protocol with a highly degenerate primer targeting coppercontaining membrane-bound monooxygenase genes for community analysis of methane-and ammonia-oxidizing bacteria[J]. FEMS Microbiol Ecol, 2017, 93(3): w244. DOI:10.1093/femsec/fiw244 |
| [15] |
HU H W, HE J Z. Comammox-a newly discovered nitrification process in the terrestrial nitrogen cycle[J]. Journal of Soils & Sediments, 2017, 17(12): 1-9. |
| [16] |
PJEVAC P, SCHAUBERGER C, POGHOSYAN L, et al. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment[J]. Frontiers in Microbiology, 2017(8): 1508. |
| [17] |
BAKER B J, LAZAR C S, TESKE A P, et al. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria[J]. Microbiome, 2015, 3(1): 3-14. DOI:10.1186/s40168-014-0064-3 |
| [18] |
MURRAY N J, MA Z, FULLER R A. Tidal flats of the Yellow Sea:A review of ecosystem status and anthropogenic threats[J]. Austral Ecology, 2015, 40(4): 472-481. DOI:10.1111/aec.2015.40.issue-4 |
| [19] |
ZHENG Y, HOU L, NEWELL S, et al. Community dynamics and activity of ammonia-oxidizing prokaryotes in intertidal sediments of the Yangtze Estuary[J]. Appl Environ Microbiol, 2013, 80(1): 408-419. |
| [20] |
STARR M P, STOLP H, TRÜPER H G, et al. The prokaryotes: a handbook on habitats, isolation and identification of bacteria[M].[S.l.]: Springer Science & Business Media, 1981.
|
| [21] |
HOU L J, LIU M, XU S Y, et al. The effects of semi-lunar spring and neap tidal change on nitrification, denitrification and N2O vertical distribution in the intertidal sediments of the Yangtze estuary, China[J]. Estuarine, Coastal and Shelf Science, 2007, 73(3/4): 607-616. |
| [22] |
ROTTHAUWE J H, WITZEL K P, LIESACK W. The ammonia monooxygenase structural gene amoa as a functional marker:Molecular fine-scale analysis of natural ammonia-oxidizing populations[J]. Appl Environ Microbiol, 1997, 63(12): 4704-4712. |
| [23] |
FRANCIS C A, ROBERTS K J, BEMAN J M, et al. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean[J]. Proc Natl Acad Sci USA, 2005, 102(41): 14683-14688. DOI:10.1073/pnas.0506625102 |
| [24] |
LI D, LIU C, LUO R, et al. MEGAHIT:An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph[J]. Bioinformatics, 2015, 31(10): 1674-1676. DOI:10.1093/bioinformatics/btv033 |
| [25] |
DR M, WALLER A, SUNAGAWA S, et al. Assessment of metagenomic assembly using simulated next generation sequencing data[J]. PLoS ONE, 2012, 7(2): e31386. DOI:10.1371/journal.pone.0031386 |
| [26] |
NIELSEN H B, ALMEIDA M, JUNCKER A S, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes[J]. Nature Biotechnology, 2014, 32(8): 822-828. DOI:10.1038/nbt.2939 |
| [27] |
KARLSSON F H, FÅK F, NOOKAEW I, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome[J]. Nature Communications, 2012, 3(4): 1245. |
| [28] |
ZELLER G, TAP J, VOIGT A Y, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer[J]. Mol Syst Biol, 2014, 10(11): 766. DOI:10.15252/msb.20145645 |
| [29] |
LI J, JIA H, CAI X, et al. An integrated catalog of reference genes in the human gut microbiome[J]. Nature Biotechnology, 2014, 32(8): 834-841. DOI:10.1038/nbt.2942 |
| [30] |
GU S, FANG L, XU X. UNIT 11.11 using SOAPaligner for short reads alignment[J]. Current Protocols in Bioinformatics, 2013, 44: 11. |
| [31] |
FU L, NIU B, ZHU Z, et al. CD-HIT:Accelerated for clustering the next-generation sequencing data[J]. Bioinformatics, 2012, 28(23): 3150-3152. DOI:10.1093/bioinformatics/bts565 |
| [32] |
COTILLARD A, KENNEDY S P, KONG L C, et al. Dietary intervention impact on gut microbial gene richness[J]. Nature, 2013, 500(7464): 585-588. DOI:10.1038/nature12480 |
| [33] |
BUCHFINK B, XIE C, HUSON D H. Fast and sensitive protein alignment using DIAMOND[J]. Nature Methods, 2014, 12(1): 59-60. |
| [34] |
QIN J, LI R, RAES J, et al. A human gut microbial gene catalogue established by metagenomic sequencing[J]. Nature, 2010, 464(7285): 59-65. DOI:10.1038/nature08821 |
| [35] |
HUSON D H, MITRA S, RUSCHEWEYH H J, et al. Integrative analysis of environmental sequences using MEGAN4[J]. Genome Research, 2011, 21(9): 1552-1560. DOI:10.1101/gr.120618.111 |
| [36] |
PRUESSE E, QUAST C, KNITTEL K, et al. SILVA:A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB[J]. Nucleic Acids Research, 2007, 35(21): 7188-7196. DOI:10.1093/nar/gkm864 |
| [37] |
GRABHERR M G, HAAS B J, YASSOUR M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome[J]. Nature Biotechnology, 2011, 29(7): 644-652. DOI:10.1038/nbt.1883 |
| [38] |
TRAPNELL C, WILLIAMS B A, PERTEA G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation[J]. Nature Biotechnology, 2010, 28(5): 511-515. DOI:10.1038/nbt.1621 |
| [39] |
LI B, DEWEY C N. RSEM:Accurate transcript quantification from RNA-Seq data with or without a reference genome[J]. BMC Bioinformatics, 2011, 12(1): 323. DOI:10.1186/1471-2105-12-323 |
| [40] |
PARK B J, PARK S J, YOON D N, et al. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria[J]. Appl Environ Microbiol, 2010, 76(22): 7575-7587. DOI:10.1128/AEM.01478-10 |
| [41] |
金文标, 李志鑫, 岳洋洋, 等.一种污水处理系统中氨氧化古细菌富集培养的方法: 中国, CN 103451120[P]. 2013-12-18. http://www.wanfangdata.com.cn/details/detail.do?_type=patent&id=CN201310041633.2
|
| [42] |
GONZALEZ-CABALEIRO R, CURTIS T P O. Study of the competition betw éen complete nitrification by a single organism and ammonia-and nitrite-oxidizing bacteria[C]//Frontiers in Wastewater Treatment and Modelling. Lecture Notes in Civil Engineering, MANNINA G, Cham, 2017: 287-291.
|
| [43] |
KITS K D, SEDLACEK C J, LEBEDEVA E V, et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle[J]. Nature, 2017, 549(7671): 269-272. DOI:10.1038/nature23679 |
| [44] |
COSTA E, PÉREZ J, KREFT J. Why is metabolic labour divided in nitrification?[J]. Trends in Microbiology, 2006, 14(5): 213-219. DOI:10.1016/j.tim.2006.03.006 |




